Master Csv File Reading In R: Ultimate Guide With Optimized Seo
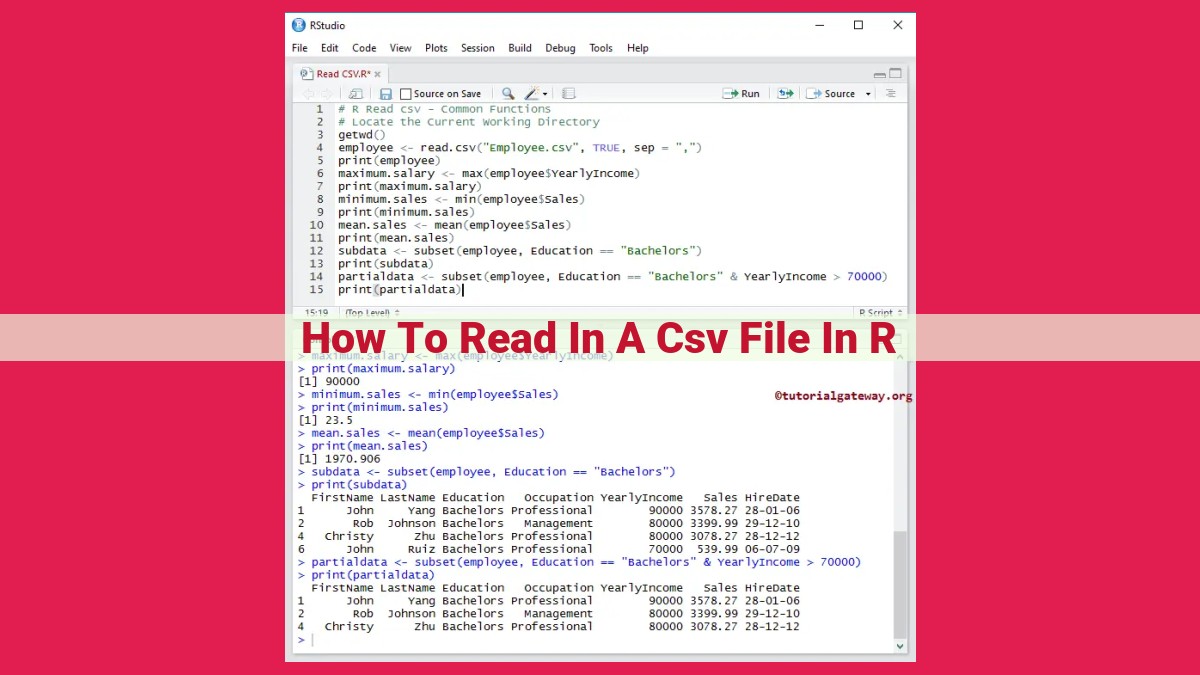
To read a comma-separated value (CSV) file in R, use the read_csv() function, specifying the file path with the file argument. Control header management, field separation, data types, handling of missing values, row/column limits, and ignore comments or leading rows with additional arguments. Ensure proper data types by specifying col_types. Handle quoted fields with quote, remove whitespaces with trim_whitespaces, and convert strings to factors with stringsAsFactors. Review real-world examples for practical applications.
Dive into the Realm of CSV Files with R: A Comprehensive Guide
In the realm of data analysis, CSV (Comma-Separated Values) files hold a significant position as a ubiquitous format for storing and exchanging data. These files organize data into a tabular structure, making them a convenient choice for various applications. As a powerful tool for data manipulation, R excels at handling CSV files, offering a comprehensive suite of functions to import, manipulate, and analyze data effectively.
R is an open-source programming language and software environment specifically designed for statistical computing and graphics. Its versatility empowers data scientists and analysts to tackle a wide range of data analysis tasks, including the management and manipulation of CSV files. By leveraging the capabilities of R, you can effortlessly import CSV data, customize its structure, and extract meaningful insights.
Unveiling the secrets of reading CSV files in R is a captivating journey that begins with the read_csv() function. This versatile function allows you to seamlessly import CSV files into R, providing a customizable framework to cater to your specific data requirements. Along this path, you’ll discover the nuances of header management, field separation, and data type specification.
Header Management plays a crucial role in organizing your data. The read_csv() function empowers you to handle missing headers, ensuring that your data remains intact and structured. Delving deeper, you’ll explore the significance of Field Separation, understanding the various separators used in CSV files and how to specify them accurately.
The ability to Specify Data Types is paramount for ensuring data integrity. R provides a range of functions, such as as.factor() and as.numeric(), to convert data into appropriate types, transforming raw data into a structured and usable format.
Dealing with Missing Values is an inevitable aspect of data analysis. R offers options for identifying and replacing missing values, enabling you to maintain data quality and prevent errors. Additionally, you’ll learn how to handle Limiting Data, Skipping Rows, and Reading Raw Data, empowering you to tailor the data import process to your specific needs.
Unveiling the secrets of Ignoring Comments and Converting Strings to Factors will further enhance your mastery of CSV file manipulation in R. You’ll discover techniques for handling comments within CSV files and explore the concept of factor data types, gaining a deeper understanding of data representation.
To solidify your understanding, the post concludes with Real-World Examples, demonstrating the practical application of the discussed concepts. These examples will showcase the power of R’s read_csv() function, providing valuable tips and suggestions to troubleshoot common issues and maximize your data analysis workflow.
Embark on this captivating journey of CSV file manipulation in R, mastering the techniques to import, customize, and analyze your data with ease. Let R be your guide as you navigate the complexities of data management, unlocking the full potential of your CSV files for insightful data analysis.
Importing CSV Data with read_csv(): A Comprehensive Guide
When working with data in R, one of the most common tasks is importing data from a Comma-Separated Values (CSV) file. CSV files are a convenient format for storing tabular data, making them easy to read and manipulate. To import CSV data into R, we use the read_csv() function, which offers a wide range of options for customizing the import process.
Introducing the read_csv() Function
The read_csv() function is part of the readr package, a powerful library for data manipulation in R. It provides an intuitive and efficient way to read CSV files, allowing you to specify various parameters to tailor the import process to your specific needs.
Handling Options for Header, Separators, and Data Types
One of the key features of read_csv() is its ability to handle different types of CSV files. You can specify whether the file has a header line using the header argument, which defaults to TRUE if the file contains a header. The sep argument allows you to specify the field separator character used in the file, which is typically a comma by default. Additionally, you can use the col_types argument to specify the data types for each column, which can be particularly useful for ensuring the correct interpretation of numeric or categorical data.
Understanding the Role of Header in CSV Files
The header line in a CSV file contains the names of the columns. By default, read_csv() expects the first line of the file to be the header and will assign these names to the columns. However, if your file does not have a header, you can set header = FALSE to read the entire file as data.
Options for Handling Missing Values in the Header
In some cases, the header line may contain missing values. To handle these situations, you can use the na.strings argument to specify a list of strings that should be interpreted as missing values in the header. For example, if you expect the header to contain the value “N/A” to indicate missing values, you can do this:
data <- read_csv("data.csv", header = TRUE, na.strings = "N/A")
Exploring Different Field Separators
CSV files can use different field separators to delimit the columns. The default separator in read_csv() is a comma, but you can specify a different separator using the sep argument. For example, if your file uses a semicolon as the separator, you can use this:
data <- read_csv("data.csv", sep = ";")
Specifying Data Types for Columns
By default, read_csv() will infer the data types for each column based on the values in the file. However, you can manually specify the data types using the col_types argument. This is especially useful if you know the expected data types beforehand or if you want to ensure consistency across different files. The col_types argument takes a vector of data types, where each element corresponds to a column in the file. For example, if you have a file with three columns: one numeric, one character, and one factor, you can specify the data types as follows:
data <- read_csv("data.csv", col_types = c("numeric", "character", "factor"))
Header Management in CSV File Reading with R
Understanding the Significance of Headers in CSV Files
Headers play a crucial role in structuring CSV (Comma-Separated Value) files. They serve as labels for each column, providing crucial information about the data contained within. Proper header management is essential for seamless data import and accurate analysis in R.
Addressing Missing Headers
In some instances, CSV files may lack explicit headers. To accommodate this, R provides the header argument in the read_csv() function. Setting header = FALSE instructs R to use the first row of the data as the header. This option is particularly useful when the file contains a blank first row or when the headers are inconsistent or missing.
Replacing Missing Header Values
Occasionally, headers may be missing or contain empty values. To handle such situations, you can use the fill argument within the read_csv() function. Simply specify the desired value (e.g., fill = "NA") to replace all missing header values. This ensures that the data frame created by read_csv() has properly labeled columns.
Optimizing Header Handling for Efficient Data Analysis
Proper header management is vital for efficient data analysis and exploration. By carefully considering the options available for header handling in read_csv(), you can ensure that your imported data is structured correctly and ready for further processing and visualization.
Field Separation in CSV Files with R’s read_csv()
When working with CSV (Comma-Separated Values) files, understanding field separation is crucial for accurate data import into R. A field separator is a character that delimits individual data values within a CSV record.
Common Separators
The most common field separator is a comma (,). However, other separators are also used, such as:
- Semicolon (;)
- Tab (\t)
- Pipe (|)
- Space ( )
Specifying Separators in read_csv()
To specify a field separator when importing a CSV file using R’s read_csv() function, use the sep argument. For example:
my_data <- read_csv("data.csv", sep = ";")
This code will import the CSV file “data.csv” using a semicolon as the field separator.
Importance of Field Separation
Correct field separation is essential for properly parsing CSV data. Incorrect separation can lead to:
- Missing or truncated data values
- Data type errors (e.g., numeric values imported as strings)
- Inaccurate data analysis and modeling
Tips for Choosing a Separator
When choosing a field separator, consider the following:
- Consistency: Ensure that the separator is consistent throughout the CSV file.
- Avoid Conflicts: Select a separator that does not appear within the data values themselves, as it may cause confusion.
- Clarity: Opt for a separator that is easily identifiable and unambiguous.
By understanding field separators and specifying them correctly in read_csv(), you can ensure that your CSV data is imported into R accurately and efficiently.
Specifying Data Types in CSV Files for Seamless Data Analytics in R
When embarking on data analysis, the importance of defining data types cannot be overstated. Accurate data types ensure the integrity of your analysis and prevent incorrect conclusions. R’s read_csv() function empowers you to specify data types for each column, ensuring that your data is interpreted correctly.
Data Type Considerations
The flexibility of CSV files allows for data of various types. However, when importing CSV data into R, you may encounter inconsistencies in data types, such as strings stored as numbers or dates represented as text. Specifying data types resolves these discrepancies, ensuring that your analysis is not compromised by incorrect data interpretation.
Data Type Conversion Functions
R provides a range of functions to convert data between different types. For instance, as.factor() converts strings to factors, while as.numeric() converts strings to numeric values. These conversion functions enable you to tailor your data to the specific requirements of your analysis.
Real-World Example
Consider a CSV file containing a column representing customer ages. Initially, these ages are stored as strings, but we need to analyze them as numeric values for statistical calculations. Using the col_types argument in read_csv(), we can specify that the “age” column should be interpreted as a numeric type:
data <- read_csv("customer_data.csv", col_types = c(age = "numeric"))
By explicitly defining the “age” column as numeric, we ensure that any subsequent statistical operations, such as calculating mean or standard deviation, will be performed correctly.
Benefits of Specifying Data Types
- Enhanced Data Accuracy: Accurate data types eliminate data interpretation errors, leading to more reliable and insightful analysis.
- Efficient Data Analysis: The correct data types allow for faster and more efficient data manipulation, as R can optimize its operations based on the specified data types.
- Improved Data Visualization: By ensuring that data types are correctly defined, you can create clear and informative data visualizations that accurately represent your findings.
Unveiling the Mystery of Missing Values: Handling Empty Cells in R with read_csv()
In the realm of data analysis, CSV files reign supreme as a versatile format for exchanging information. However, one common challenge encountered when working with CSV files is dealing with missing values. These empty cells can wreak havoc on your analysis, but fear not! R, the wizard of data manipulation, offers a spellbinding array of options to help you identify, replace, and remove missing values using the read_csv() function.
Identifying Missing Values: A Quest for Empty Cells
The first step in tackling missing values is to identify them. R provides two handy functions for this task: is.na() and is.null(). These functions return TRUE for missing values and FALSE otherwise. You can use them to create a mask indicating the location of missing values in your data frame.
Replacing Missing Values: Filling the Void
Once you’ve identified the missing values, it’s time to replace them. The na.strings argument in read_csv() allows you to specify values that should be treated as missing. For example, you can replace all empty cells with NA (Not Applicable) or a custom placeholder like -999.
Removing Missing Values: A Clean Sweep
If you prefer to remove missing values entirely, R has got you covered. The na.rm argument in read_csv() allows you to ignore rows or columns containing missing values. This can be useful if you’re confident that the missing data won’t significantly impact your analysis.
By harnessing the power of R’s read_csv() function, you can effortlessly identify, replace, or remove missing values from your CSV files. This empowers you to ensure the accuracy and reliability of your data analysis, leading you to insightful discoveries and a deeper understanding of your data.
Limiting Data: Efficiently Importing Large Datasets
In the realm of data analysis, dealing with large datasets is a common challenge. Importing every single row of data can be time-consuming and unnecessary. Limiting data import offers a solution, allowing you to focus on a subset of your data, saving time and resources.
Why Limit Data?
- Performance optimization: Importing a smaller subset of data significantly improves performance, reducing loading and processing times.
- Focused analysis: By limiting the data to a specific timeframe or group, you can target your analysis, ensuring that you’re examining the most relevant information.
- Resource conservation: Memory and storage limitations may necessitate limiting data import, especially when dealing with massive datasets.
Getting Row and Column Counts
Before limiting data, it’s essential to understand the size of your dataset. The ncol() and nrow() functions provide quick insights:
ncol()returns the number of columns in your data frame.nrow()returns the number of rows in your data frame.
Limiting Data with nrows
The nrows argument in the read_csv() function allows you to specify the number of rows to import. It’s useful when you want to:
- Preview a small portion of data for quick insights.
- Sample a large dataset for exploratory analysis.
- Break down data import into manageable chunks for processing.
For instance, to import the first 100 rows of a CSV file called “data.csv”:
data <- read_csv("data.csv", nrows = 100)
Additional Benefits
Limiting data import not only provides better performance but also opens up other possibilities:
- Incremental data loading: You can import data in batches, providing real-time updates to your analysis.
- Random sampling: Limiting data via
nrowsallows for random sampling, ensuring that you obtain a representative sample of your dataset. - Troubleshooting: Isolating issues in data import can be easier when working with a smaller subset of data.
By understanding the capabilities of nrows, you can optimize your data import process, saving time and effort while maximizing the value of your analysis.
Skipping Rows (skip)
- Ignoring unwanted rows at the start of the file
Skipping Rows: Ignoring Unwanted Lines in CSV Files with R’s read_csv()
Imagine you’re working with a CSV file that contains valuable data but is cluttered with unnecessary information at the beginning. You could manually remove these rows, but there’s a better way: using R’s read_csv() function with the skip argument.
The skip argument allows you to specify the number of rows to ignore at the start of your CSV file. This is incredibly useful when dealing with files that have header rows, comments, or other non-data content that you don’t need.
To use skip, simply pass the number of rows you want to omit as an argument to the read_csv() function. For example, if you have a CSV file with three header rows, you would use the following code:
data <- read_csv("my_csv_file.csv", skip = 3)
This will create a data frame that contains only the data from the fourth row onward.
Benefits of Skipping Rows
Skipping rows can save you time and improve the efficiency of your data analysis. By removing unwanted lines, you can:
- Reduce the size of your data frame, making it easier to work with.
- Focus on the relevant data without having to manually filter out unwanted rows.
- Avoid errors caused by incorrect data types or formatting issues in the skipped rows.
Tips for Using skip
- Determine the number of rows to skip by examining the header or viewing the first few rows of your CSV file.
- Use the
head()function to preview the data and ensure that you’re skipping the correct number of rows. - If you’re unsure about the number of rows to skip, start with a small value and gradually increase it until you reach the desired result.
Dive into Raw Data with R: A Guide to Importing CSV Files as Text
In the realm of data analysis, CSV files reign supreme as a simple yet powerful way to store and exchange data. R, a programming language tailored for data manipulation, offers an array of options for importing CSV data, including the ability to read raw text.
When dealing with CSV files, we can encounter various data formats and structures. To accommodate these variations, R provides the as.is parameter in the read_csv() function. This parameter allows us to import data as raw text, giving us complete control over how the data is parsed and formatted.
Unveiling Raw Data
Importing data as raw text grants us the flexibility to mold the data to our specific needs. R offers two functions for creating data structures from raw text: data.frame() and matrix().
data.frame() creates a data frame, which is a tabular structure with rows and columns, while matrix() creates a matrix, which is a rectangular arrangement of data elements.
Example in Action
Let’s consider a CSV file containing raw data, such as:
age,name,city
21,Alice,New York
25,Bob,London
To import this data as raw text, we can use the following R code:
raw_data <- read_csv("raw_data.csv", as.is = TRUE)
This code assigns the raw data to the raw_data variable. We can then use the data.frame() or matrix() function to create a data frame or matrix from the raw data.
Creating a Data Frame from Raw Data
To create a data frame from the raw data, we can use the following code:
data_frame <- data.frame(raw_data)
The data.frame() function takes the raw data as an argument and creates a data frame with the following structure:
| age | name | city |
|---|---|---|
| 21 | Alice | New York |
| 25 | Bob | London |
Creating a Matrix from Raw Data
To create a matrix from the raw data, we can use the following code:
matrix <- matrix(raw_data, nrow = 2, byrow = TRUE)
The matrix() function takes the raw data and the number of rows (nrow) as arguments. The byrow parameter specifies whether the data should be filled in by row or by column. In this case, we specify byrow = TRUE to fill in the matrix by row.
The resulting matrix will have the following structure:
[,1] [,2] [,3]
[1,] "age" "name" "city"
[2,] "21" "Alice" "New York"
Importing CSV data as raw text in R provides us with the power to manipulate data to our specific requirements. By utilizing the as.is parameter in read_csv(), we can read raw data and transform it into data frames or matrices using data.frame() and matrix(), respectively. This flexibility empowers us to handle a wide range of data formats and structures, unlocking the full potential of our data analysis in R.
Ignore the Noise: Handling Comments in CSV Files with R’s read_csv()
When working with CSV (Comma-Separated Value) files, we often encounter comments that provide additional information or пояснения. These comments, though helpful for understanding the data, can sometimes interfere with the import process. R’s read_csv() function provides a convenient solution for this by allowing us to specify a character that marks the beginning of a comment.
To specify a comment character, we use the comment argument in read_csv(). By default, it’s set to NA, which means no character is treated as a comment. However, we can change it to any character we want. For example, if our comments start with the pound sign (#), we would use comment = "#".
Here’s a simple illustration:
# Read CSV file with comments starting with "#"
data <- read_csv("data.csv", comment = "#")
In this example, the data.csv file contains comments that begin with the “#” character. By specifying comment = "#" in read_csv(), we instruct R to ignore all lines that start with this character.
This can be particularly useful when working with large datasets, where comments may introduce unnecessary noise and make data processing more challenging. By ignoring comments, we ensure that only the relevant data is imported into our R environment.
Specifying comment characters in read_csv() is a simple yet effective way to handle comments in CSV files. It allows us to cleanse our data and focus on the information that truly matters for our analysis.
Converting Strings to Factors (stringsAsFactors)
- Understanding factor data type
- Converting strings to factors (as.factor(), factor())
Converting Strings to Factors: Unlocking the Power of Categorical Data
When working with data in R, it’s essential to understand the different data types and their appropriate representation. One crucial aspect of data manipulation is converting strings to factors. Factors are a special data type in R that represents categorical variables, which are non-numeric and have a finite number of distinct values.
The Essence of Factors
Think of factors as a way to organize and label categorical data. For instance, imagine a dataset with a column titled “gender” containing values like “male,” “female,” and “unknown.” Converting this column to a factor allows R to recognize these values as categories rather than simple text strings.
Transforming Strings to Factors
Converting strings to factors is straightforward. You can use the as.factor() function or the factor() function. Here’s an example using as.factor():
gender_factor <- as.factor(gender)
This function converts the “gender” column, which originally contained strings, into a factor variable.
Unveiling the Power of Factors
Factors offer several advantages for data analysis. They allow for:
- Categorical Analysis: Factors facilitate statistical operations specifically designed for categorical data, such as frequency tables and chi-square tests.
- Improved Visualization: Factors enable the creation of insightful visualizations, such as bar charts and box plots, that highlight the distribution of categories.
- Efficient Handling of Missing Values: Factors handle missing values gracefully, replacing them with a dedicated “NA” category.
Examples in Action
Let’s illustrate the usage of as.factor() with a real-world example. Suppose you have a dataset of customer purchase records with a column called “product_type.” To analyze the distribution of products sold, you can convert this column to a factor:
product_type_factor <- as.factor(product_type)
This conversion will create a factor variable with each unique product type as a category. You can then use this factor variable in statistical tests or visualizations to gain insights into the popularity of different products.
Converting strings to factors is a fundamental data manipulation technique in R. By understanding the concept of factors and utilizing the as.factor() function effectively, you can unlock the power of categorical data analysis. Remember, using the correct data type ensures accurate and meaningful outcomes in your statistical endeavors.
Understanding the Decimal Point Character in CSV Files
When working with CSV files, it’s crucial to be aware of the different decimal point characters used to represent fractional values. These characters vary depending on the region and locale of the data source, and they can cause confusion if not handled correctly.
In many countries, the dot (.) is used as the decimal separator, while in other regions, such as Europe and South America, the comma (,) serves this purpose. This distinction is important because R assumes the dot as the default decimal separator when reading CSV files.
Consider the following example:
value
1.2345
1,2345
If this CSV file is imported into R without specifying the decimal point character, the first value will be interpreted correctly as 1.2345, while the second value will be read as 12,345 due to the presence of the comma.
Specifying the Decimal Point Character
To ensure accurate data import, it’s essential to specify the correct decimal point character using the dec argument in the read_csv() function. This argument allows you to set the character that should be treated as the decimal separator.
data <- read_csv("data.csv", dec = ",")
By setting dec = ",", R will expect commas to separate the fractional part of values in the CSV file, ensuring that the second value in the example above is correctly imported as 1.2345.
Troubleshooting Decimal Point Character Issues
Incorrectly specifying the decimal point character can lead to data inaccuracies. If you encounter unexpected results when importing CSV data, consider checking the following:
- The locale settings of your R installation
- The expected format of the decimal point character in the CSV file
- The
decargument in theread_csv()function call
Understanding the decimal point character in CSV files is essential for accurate data import in R. By specifying the correct decimal separator using the dec argument, you can ensure that your data is interpreted as intended and avoid potential errors or data loss.
Field Quoting in CSV Files: A Guide for Seamless Data Import
CSV (Comma-Separated Values) files are a common format for data exchange. Understanding how to handle field quoting ensures accurate and efficient data import in R.
What is Field Quoting?
In CSV files, fields (data values) can be enclosed in characters like double or single quotes. This quoting helps distinguish fields containing special characters (e.g., commas, spaces) from the delimiters (e.g., commas) that separate fields.
Quoting Options in R
The read_csv() function in R provides the quote parameter to specify how fields should be quoted during import.
- “ (Default): Double quotes enclose all fields, ensuring that all data is treated as text.
- “”: No quoting; all fields are considered as raw text, and special characters may cause errors.
- NULL: Quotes are ignored; all fields are treated as raw text.
Choosing the Right Quoting Option
The appropriate quoting option depends on the specific data file. If fields contain special characters or spaces that could be misinterpreted as delimiters, using double quotes (“”) is recommended.
Examples
Consider the following CSV file:
"John Doe", "2023-03-08", 100
"Jane Smith", "2023-03-10", 200
"Bob Jones", "2023-03-12", 300
If we import this file without specifying quoting, we might face errors due to the spaces in names and commas in dates.
data <- read_csv("data.csv")
To ensure accurate import, we can use double quotes:
data <- read_csv("data.csv", quote = '"')
Field quoting plays a crucial role in importing CSV data into R. Understanding the various quoting options and their impact on data interpretation enables you to handle CSV files efficiently and accurately, ensuring reliable data analysis.
Trimming White Spaces: Declutter Your CSV Data for Clarity
Whitespace Overload:
CSV files can often be cluttered with unnecessary white spaces—spaces, tabs, or line breaks that creep in between data elements. These unwanted characters can create inconsistencies and hinder data analysis.
Benefits of Decluttering:
Trimming white spaces ensures consistent data formatting, making it easier to:
– Identify patterns and trends
– Merge datasets without data conflicts
– Improve data quality for machine learning and visualizations
Whitespace Removal Techniques:
R provides several functions for whitespace removal:
- gsub(): A powerful string substitution function that can replace white spaces with empty strings.
- str_trim(): Specifically designed for whitespace trimming, it removes leading and trailing white spaces.
Code Snippets:
To remove all white spaces from a CSV column named “Name”:
df$Name <- gsub(" ", "", df$Name)
To remove only leading and trailing white spaces:
df$Name <- str_trim(df$Name)
Real-World Impact:
Whitespace trimming is crucial for:
– Data cleaning: Preparing data for analysis by removing inconsistencies.
– Data integration: Combining datasets from different sources with varied formatting.
– Data standardization: Ensuring consistent data formats for effective data sharing and collaboration.
Mastering CSV File Reading in R
In the realm of data analysis, CSV (Comma-Separated Values) files reign supreme. These versatile formats store data in a structured, text-based manner, making them easily accessible by various applications. If you’re a data enthusiast or R user, this comprehensive guide will equip you with the essential knowledge and techniques for reading CSV files in R.
Kick-off with read_csv()
read_csv() is the gateway to importing CSV data into R. This powerful function offers a plethora of options to customize your import process. From handling headers and separators to specifying data types and filling missing values, you’ll master the art of data manipulation in no time.
Header Harmony (header)
Headers play a crucial role in CSV files, identifying each column’s purpose. read_csv() lets you fine-tune your header management. You can skip headers altogether (if they’re not present), specify a row number to use as the header, or even replace missing headers with custom names.
Field Separation: A Delicate Balance (sep)
CSV files rely on specific characters to separate fields, and read_csv() gives you the flexibility to accommodate these variations. Whether it’s a comma, semicolon, tab, or other delimiter, you can tailor your import process to match the format of your data.
Unleashing Data Types (col_types)
Data types are essential for accurate data analysis. read_csv() provides an array of functions (e.g., as.factor(), as.numeric()) to convert your imported data into specific types. This ensures that R interprets your data correctly, preventing errors and misinterpretations.
Handling Missing Values with Grace (na.strings)
Missing values are a common challenge in data handling. read_csv() arms you with options to identify and replace or remove these values. You can specify specific strings to represent missing values and opt to remove entire rows or columns if they contain missing data.
Real-World Explorations
Now, let’s dive into practical examples of how to use read_csv() with different options. We’ll explore scenarios such as limiting data import, skipping unwanted rows, importing raw data, and managing comments. Along the way, we’ll share tips and tricks for troubleshooting common issues, ensuring you become a CSV-reading pro in R.